Selected publications
For a full list please visit Google Scholar
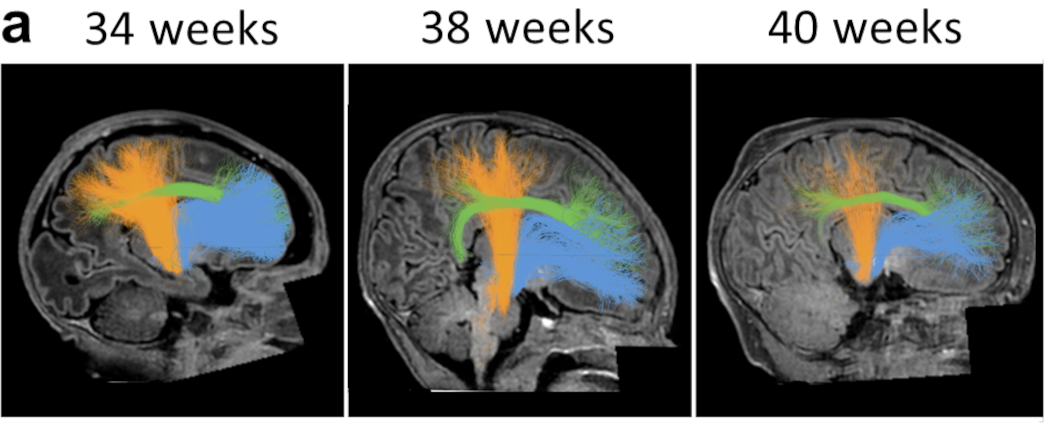
The formation of myelin, the fatty sheath that insulates nerve fibers, is critical for healthy brain function. A fundamental open question is what impact being born has on myelin growth. To address this, we evaluated a large (n=300) cross-sectional sample of newborns from the Developing Human Connectome Project (dHCP). First, we developed new software for the automated identification of 20 white matter bundles in individuals that is well-suited for large samples. Next, we fit linear models that quantify how T1w/T2w (a myelin-sensitive imaging contrast) changes over time at each point along the bundles. We found faster growth of T1w/T2w along the lengths of all bundles before birth than right after birth. Further, in a separate longitudinal sample of preterm infants (N=34), we found lower T1w/T2w than in full-term peers measured at the same age. By applying the linear models fit on the cross-section sample to the longitudinal sample of preterm infants, we find that their delay in T1w/T2w growth is well explained by the amount of time they spent developing in utero and ex utero. These results suggest that white matter myelinates faster in utero than ex utero . The reduced rate of myelin growth after birth, in turn, explains lower myelin content in individuals born preterm, and could account for long-term cognitive, neurological, and developmental consequences of preterm birth. We hypothesize that closely matching the environment of infants born preterm to what they would have experienced in the womb may reduce delays in myelin growth and hence improve developmental outcomes.
Mareike Grotheer, David Bloom, John Kruper, Adam Richie-Halford, Stephanie Zika, Vicente A. Aguilera González, Jason D. Yeatman, Kalanit Grill-Spector & Ariel Rokem
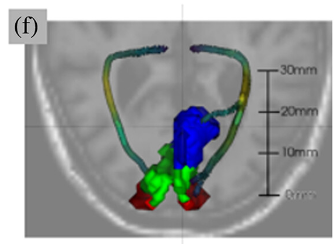
The neural pathways that carry information from the foveal, macular, and peripheral visual fields have distinct biological properties. The optic radiations (OR) carry foveal and peripheral information from the thalamus to the primary visual cortex (V1) through adjacent but separate pathways in the white matter. Here, we perform white matter tractometry using pyAFQ on a large sample of diffusion MRI (dMRI) data from subjects with healthy vision in the U.K. Biobank dataset (UKBB; N = 5,382; age 45–81). We use pyAFQ to characterize white matter tissue properties in parts of the OR that transmit information about the foveal, macular, and peripheral visual fields, and to characterize the changes in these tissue properties with age. We find that (1) independent of age there is higher fractional anisotropy, lower mean diffusivity, and higher mean kurtosis in the foveal and macular OR than in peripheral OR, consistent with denser, more organized nerve fiber populations in foveal/parafoveal pathways, and (2) age is associated with increased diffusivity and decreased anisotropy and kurtosis, consistent with decreased density and tissue organization with aging. However, anisotropy in foveal OR decreases faster with age than in peripheral OR, while diffusivity increases faster in peripheral OR, suggesting foveal/peri-foveal OR and peripheral OR differ in how they age.
John Kruper, Noah C. Benson, Sendy Caffarra, Julia Owen, Yue Wu, Aaron Y. Lee, Cecilia S. Lee, Jason D. Yeatman, Ariel Rokem & UK Biobank Eye and Vision Consortium
Human Brain Mapping 44 (8), 3123-3135
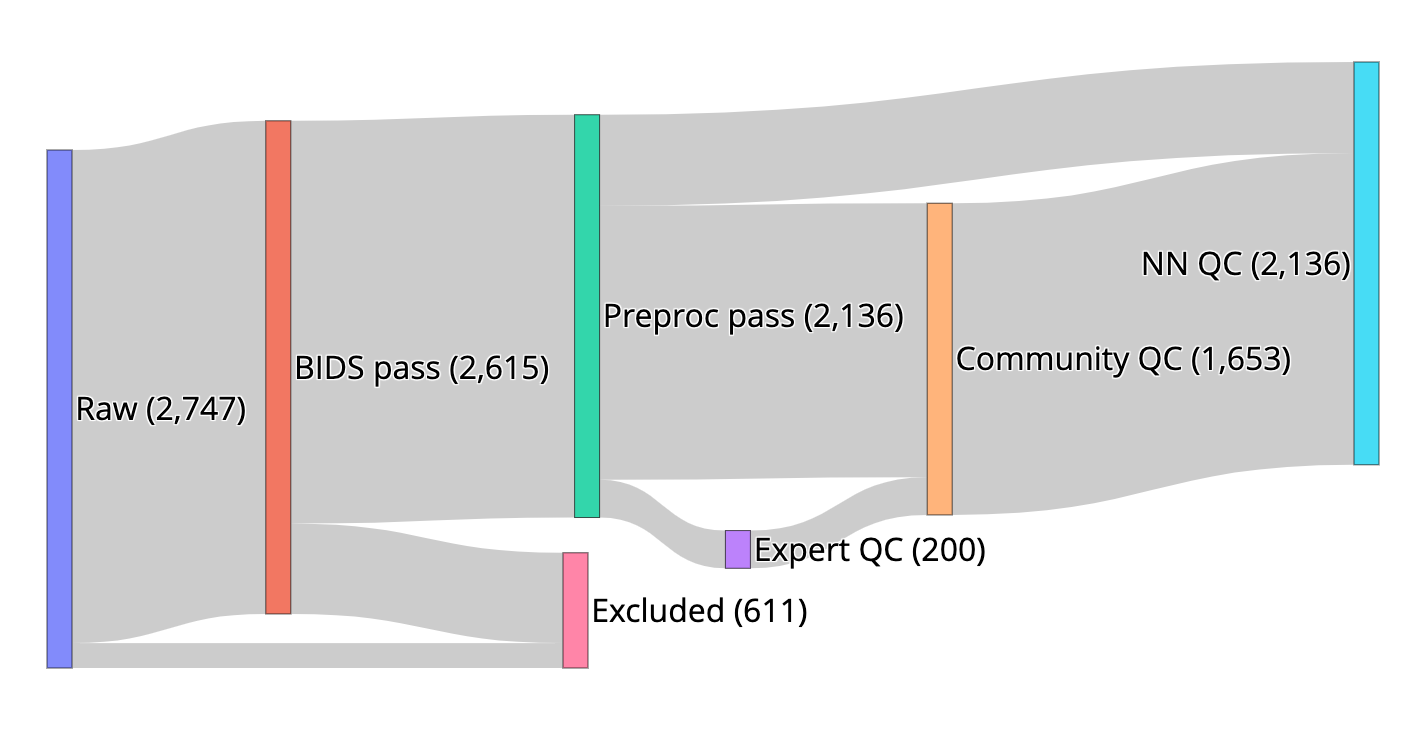
We created resources to facilitate research on the role of human brain microstructure in the development of mental health disorders, based on openly-available diffusion MRI (dMRI) data from the Healthy Brain Network (HBN) study. First, we curated the HBN dMRI data (N=2,747) into the Brain Imaging Data Structure and preprocessed it according to best-practices, including denoising and correcting for motion effects, susceptibility-related distortions, and eddy currents. Preprocessed, analysis-ready data was made openly available. Data quality plays a key role in the analysis of dMRI, and we provide automated quality control (QC) scores for every scan, as part of the data release. To scale QC to this large dataset, we trained a neural network through the combination of a small data subset scored by experts and a larger set scored by community scientists. The network performs QC highly concordant with that of experts on a held out set (ROC-AUC = 0.947). A further analysis of the neural network demonstrates that it relies on image features with relevance to QC. Altogether, this work both delivers a resource for transdiagnostic research in brain connectivity and pediatric mental health and serves as a novel tool for automated QC of large datasets.
Adam Richie-Halford, Matthew Cieslak, Lei Ai, Sendy Caffarra, Sydney Covitz, Alexandre R. Franco, Iliana I. Karipidis, John Kruper, Michael Milham, Bárbara Avelar-Pereira, Ethan Roy, Valerie J. Sydnor, Jason Yeatman, The Fibr Community Science Consortium, Theodore D. Satterthwaite, Ariel Rokem
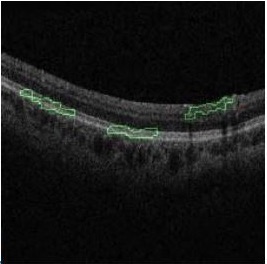
Glaucoma, the leading cause of irreversible blindness worldwide, is a disease that damages the optic nerve. Current machine learning (ML) approaches for glaucoma detection rely on features such as retinal thickness maps; however, the high rate of segmentation errors when creating these maps increase the likelihood of faulty diagnoses. This paper proposes a new, comprehensive, and more accurate ML-based approach for population-level glaucoma screening. Our contributions include: (1) a multi-modal model built upon a large data set that includes demographic, systemic and ocular data as well as raw image data taken from color fundus photos (CFPs) and macular Optical Coherence Tomography (OCT) scans, (2) model interpretation to identify and explain data features that lead to accurate model performance, and (3) model validation via comparison of model output with clinician interpretation of CFPs. We also validated the model on a cohort that was not diagnosed with glaucoma at the time of imaging but eventually received a glaucoma diagnosis. Results show that our model is highly accurate (AUC 0.97) and interpretable. It validated biological features known to be related to the disease, such as age, intraocular pressure and optic disc morphology. Our model also points to previously unknown or disputed features, such as pulmonary capacity and retinal outer layers.
Parmita Mehta, Christine A Petersen, Joanne C Wen, Michael R Banitt, Philip P Chen, Karine D Bojikian, Catherine Egan, Su-In Lee, Magdalena Balazinska, Aaron Y Lee & Ariel Rokem (last two authors contributed equally)
American Journal of Ophthalmology 231:154-169
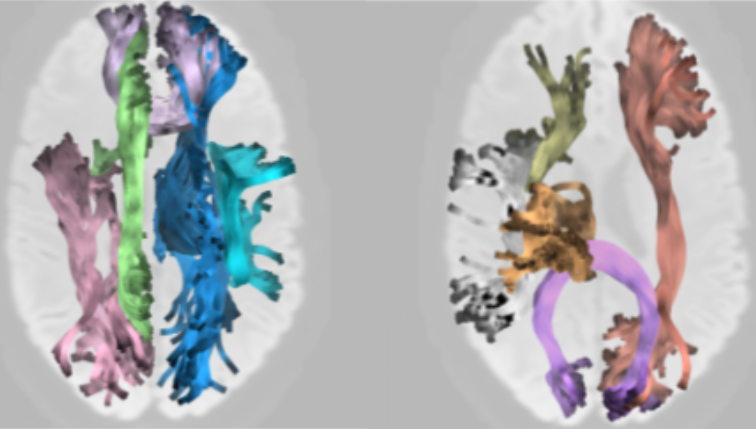
The validity of research results depends on the reliability of analysis methods. In recent years, there have been concerns about the validity of research that uses diffusion-weighted MRI (dMRI) to understand human brain white matter connections in vivo, in part based on reliability of the analysis methods used in this field. We defined and assessed three dimensions of reliability in dMRI-based tractometry, an analysis technique that assesses the physical properties of white matter pathways: (1) reproducibility, (2) test-retest reliability and (3) robustness. To facilitate reproducibility, we provide software that automates tractometry (https://yeatmanlab.github.io/pyAFQ). In measurements from the Human Connectome Project, as well as clinical-grade measurements, we find that tractometry has high test-retest reliability that is comparable to most standardized clinical assessment tools. We find that tractometry is also robust: showing high reliability with different choices of analysis algorithms. Taken together, our results suggest that tractometry is a reliable approach to analysis of white matter connections. The overall approach taken here both demonstrates the specific trustworthiness of tractometry analysis and outlines what researchers can do to demonstrate the reliability of computational analysis pipelines in neuroimaging.
John Kruper, Jason Yeatman, Adam Richie-Halford, David Bloom, Mareike Grotheer, Sendy Caffarra, Gregory Kiar, Iliana Karipidis, Ethan Roy & Ariel Rokem
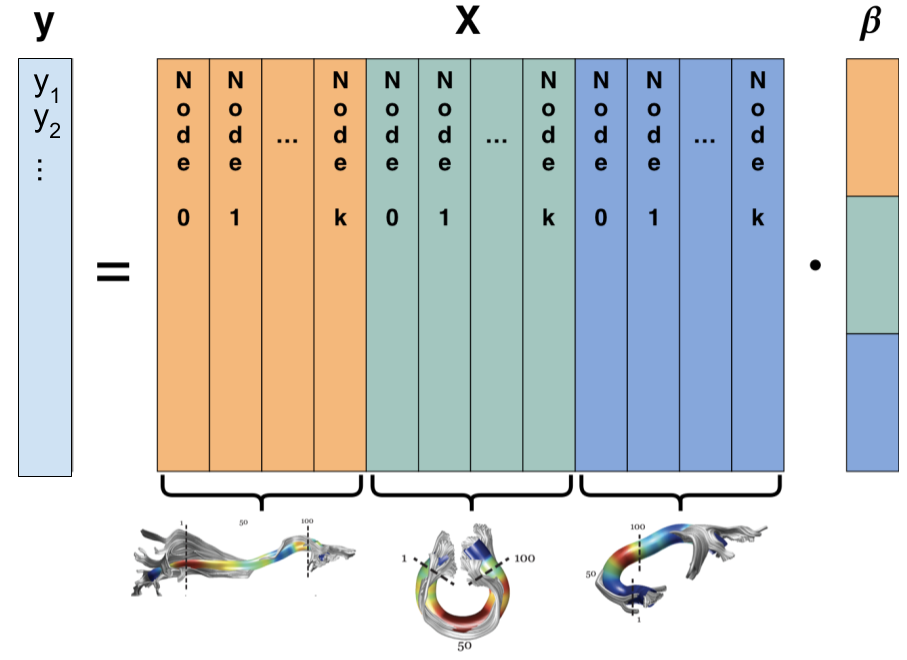
The white matter contains long-range connections between different brain regions and the organization of these connections holds important implications for brain function in health and disease. Tractometry uses diffusion-weighted magnetic resonance imaging (dMRI) data to quantify tissue properties along the trajectories of these connections. In the present work, we developed a method based on the sparse group lasso (SGL) that takes into account tissue properties measured along all of the bundles, and selects informative features by enforcing sparsity, not only at the level of individual bundles, but also across the entire set of bundles and all of the measured tissue properties. The sparsity penalties for each of these constraints is identified using a nested cross-validation scheme that guards against over-fitting and simultaneously identifies the correct level of sparsity. SGL makes it possible to leverage the multivariate relationship between diffusion properties measured along multiple bundles to make accurate predictions of subject characteristics while simultaneously discovering the most relevant features of the white matter for the characteristic of interest.
Adam Richie-Halford, Jason Yeatman, Noah Simon & Ariel Rokem
PLoS Computational Biology: 17(6): e1009136
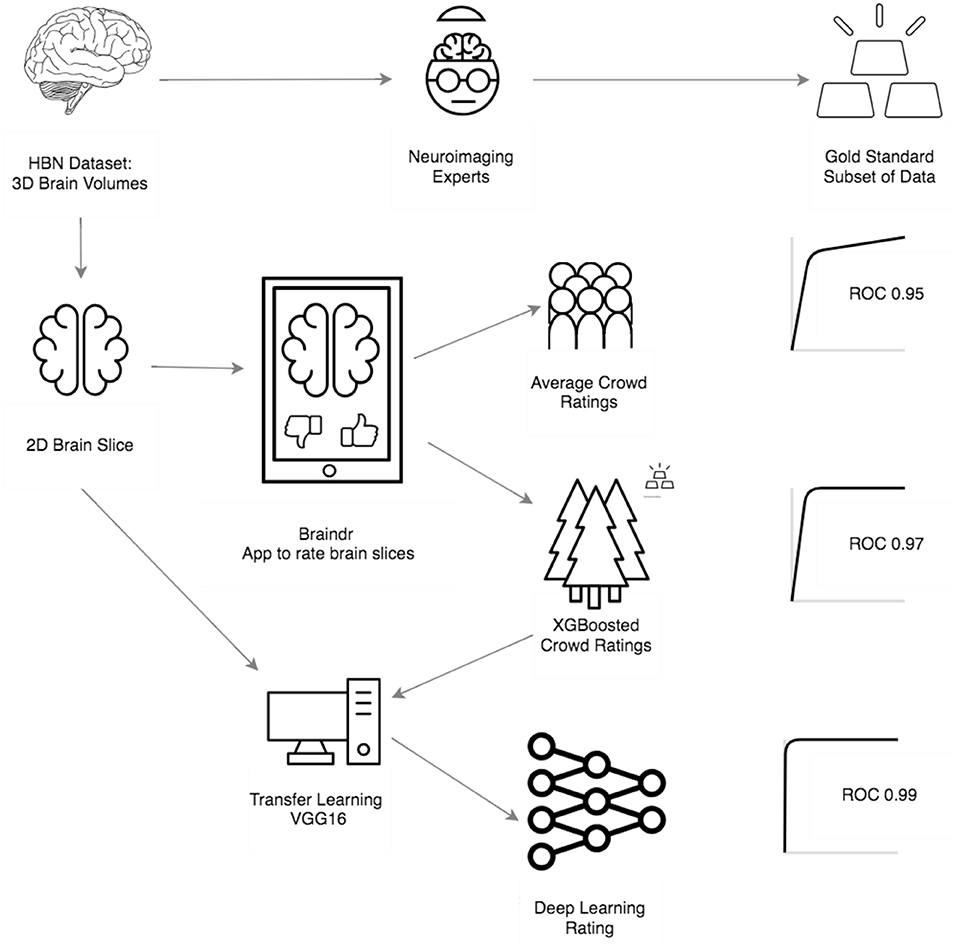
Combining citizen science and deep learning can generalize and scale expert decision making; this is particularly important in disciplines where specialized, automated tools do not yet exist. In Braindr, expert-labeled data were amplified by citizen scientists through a simple web interface. A deep learning algorithm was then trained to predict data quality, based on citizen scientist labels. Deep learning performed as well as specialized algorithms for quality control (AUC = 0.99).
Anisha Keshavan, Jason Yeatman & Ariel Rokem
Frontiers in Neuroinformatics, 13: 29
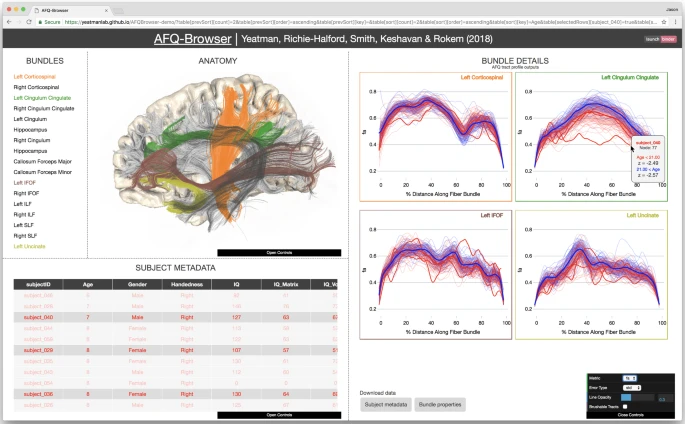
Human neuroscience research faces several challenges with regards to reproducibility. While scientists are generally aware that data sharing is important, it is not always clear how to share data in a manner that allows other labs to understand and reproduce published findings. Here we report a new open source tool, AFQ-Browser, that builds an interactive website as a companion to a diffusion MRI study. Because AFQ-Browser is portable—it runs in any web-browser—it can facilitate transparency and data sharing. Moreover, by leveraging new web-visualization technologies to create linked views between different dimensions of the dataset (anatomy, diffusion metrics, subject metadata), AFQ-Browser facilitates exploratory data analysis, fueling new discoveries based on previously published datasets. In an era where Big Data is playing an increasingly prominent role in scientific discovery, so will browser-based tools for exploring high-dimensional datasets, communicating scientific discoveries, aggregating data across labs, and publishing data alongside manuscripts.
Jason Yeatman, Adam Richie-Halford, Josh Smith, Anisha Keshavan & Ariel Rokem
Nature Communications: 9, Article number: 940
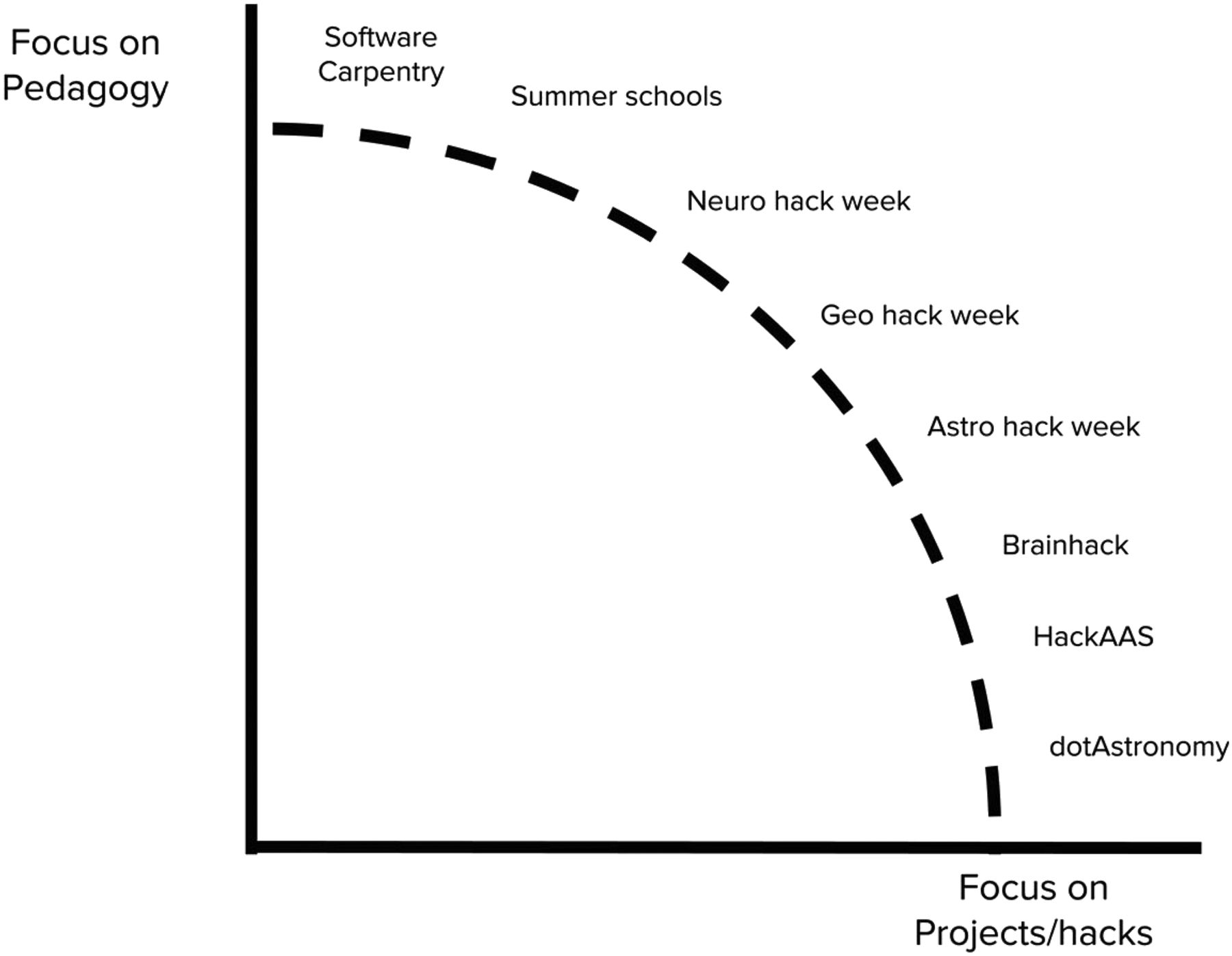
As scientific disciplines grapple with more datasets of rapidly increasing complexity and size, new approaches are urgently required to introduce new statistical and computational tools into research communities and improve the cross-disciplinary exchange of ideas. In this paper, we introduce a type of scientific workshop, called a hack week, which allows for fast dissemination of new methodologies into scientific communities and fosters exchange and collaboration within and between disciplines. We present implementations of this concept in astronomy, neuroscience, and geoscience and show that hack weeks produce positive learning outcomes, foster lasting collaborations, yield scientific results, and promote positive attitudes toward open science.
Daniela Huppenkothen, Anthony Arendt, David W. Hogg, Karthik Ram, Jacob T. VanderPlas, & Ariel Rokem